
Caret, nproc and naivebayes
Source:vignettes/caret, nproc and naivebayes.Rmd
caret, nproc and naivebayes.RmdIntroduction
The naive_bayes() function, a powerful tool for
classification, is readily accessible through the highly regarded Caret
package. Additionally, it can be utilized seamlessly via the nproc package,
providing users with alternative options. This concise documentation
aims to demonstrate the basic usage of the naive_bayes()
function in both the Caret
and nproc
packages.
Naive Bayes in Caret
The first part showcases how to train a Naive Bayes model using the
naive_bayes() function within the caret
interface in R. It provides an example of how to prepare the data, train
the Naive Bayes model, and perform classification using the trained
model. This section focuses on the core steps of training a Naive Bayes
model and utilizing it for classification tasks.
The second part demonstrates the process of defining a tuning grid,
performing resampling, and finding the “optimal” Naive Bayes model using
the caret package in R. It outlines how to define a grid of
tuning parameters, train multiple models with different parameter
combinations, and select the best-performing model based on resampling
techniques. This section emphasizes the importance of tuning the Naive
Bayes model to achieve better performance and provides insights into the
parameter selection process.
By combining these two sections, users can gain a comprehensive
understanding of how to train and optimize Naive Bayes models using the
naive_bayes() function within the caret
interface in R.
Model Training with Caret
The model is trained using the caret::train() function,
specifying "naive_bayes" as the method and setting
usepoisson to TRUE to handle the count variable.
After training the model, classification can be performed by using the trained model to predict the class labels. Additionally, the model provides posterior probabilities, which can be obtained for further analysis.
If needed, the code allows accessing the underlying
naive_bayes object, providing access to additional
information and functionalities of the Naive Bayes model.
Overall, the code demonstrates the process of training a Naive Bayes
model, performing classification, and accessing the model object for
further analysis using the caret interface in R.
library(caret, quietly = TRUE)
library(naivebayes)## naivebayes 1.0.0 loaded## For more information please visit:## https://majkamichal.github.io/naivebayes/
# Load the iris dataset
data(iris)
# Prepare the data
new <- iris[-c(1,2,3)]
set.seed(1)
new$Discrete <- sample(LETTERS[1:3], nrow(new), TRUE)
set.seed(1)
new$Counts <- c(rpois(50, 1), rpois(50, 2), rpois(50, 10))
# Train the Naive Bayes model with the Caret package
naive_bayes_via_caret <- train(Species ~ .,
data = new,
method = "naive_bayes",
usepoisson = TRUE)
## Print the trained model
# naive_bayes_via_caret
# Perform classification
head(predict(naive_bayes_via_caret, newdata = new))## [1] setosa setosa setosa setosa setosa setosa
## Levels: setosa versicolor virginica## setosa versicolor virginica
## 1 1.0000000 2.004949e-08 9.549385e-14
## 2 1.0000000 4.242500e-08 3.229798e-13
## 3 1.0000000 2.512936e-08 1.566929e-13
## 4 0.9999999 5.203335e-08 5.710476e-13
## 5 1.0000000 2.004949e-08 9.549385e-14
## 6 0.9999514 4.860925e-05 4.128918e-10
# Access the underlying naive_bayes object
nb_object <- naive_bayes_via_caret$finalModel
class(nb_object)## [1] "naive_bayes"
# nb_objectParameter Tuning with Caret
First, a tuning grid is defined using the expand.grid()
function, which specifies different combinations of tuning parameters
for the Naive Bayes model.
Then, the Naive Bayes model is fitted with parameter tuning using the
caret::train() function. The tuneGrid argument
is used to provide the defined tuning grid. The resulting model is
stored in the naive_bayes_via_caret2 object.
To view the selected tuning parameters, you can access
naive_bayes_via_caret2$finalModel$tuneValue. If you
uncomment the respective line of code, you can also view the final Naive
Bayes model itself.
The tuning process can be visualized using the generic
plot() function, which provides insights into the
performance of different parameter combinations.
Finally, the trained model is used to perform classification on new
data by using the predict() function. The class labels of
the new data can be obtained with
predict(naive_bayes_via_caret2, newdata = new).
# Define tuning grid
nb_grid <- expand.grid(usekernel = c(TRUE, FALSE),
laplace = c(0, 0.5, 1),
adjust = c(0.75, 1, 1.25, 1.5))
# Fit the Naive Bayes model with parameter tuning
set.seed(2550)
naive_bayes_via_caret2 <- train(Species ~ .,
data = new,
method = "naive_bayes",
usepoisson = TRUE,
tuneGrid = nb_grid)
# View the selected tuning parameters
naive_bayes_via_caret2$finalModel$tuneValue## laplace usekernel adjust
## 16 0 TRUE 1.5
## View the final naive_bayes model
# naive_bayes_via_caret2$finalModel
# Visualize the tuning process
plot(naive_bayes_via_caret2)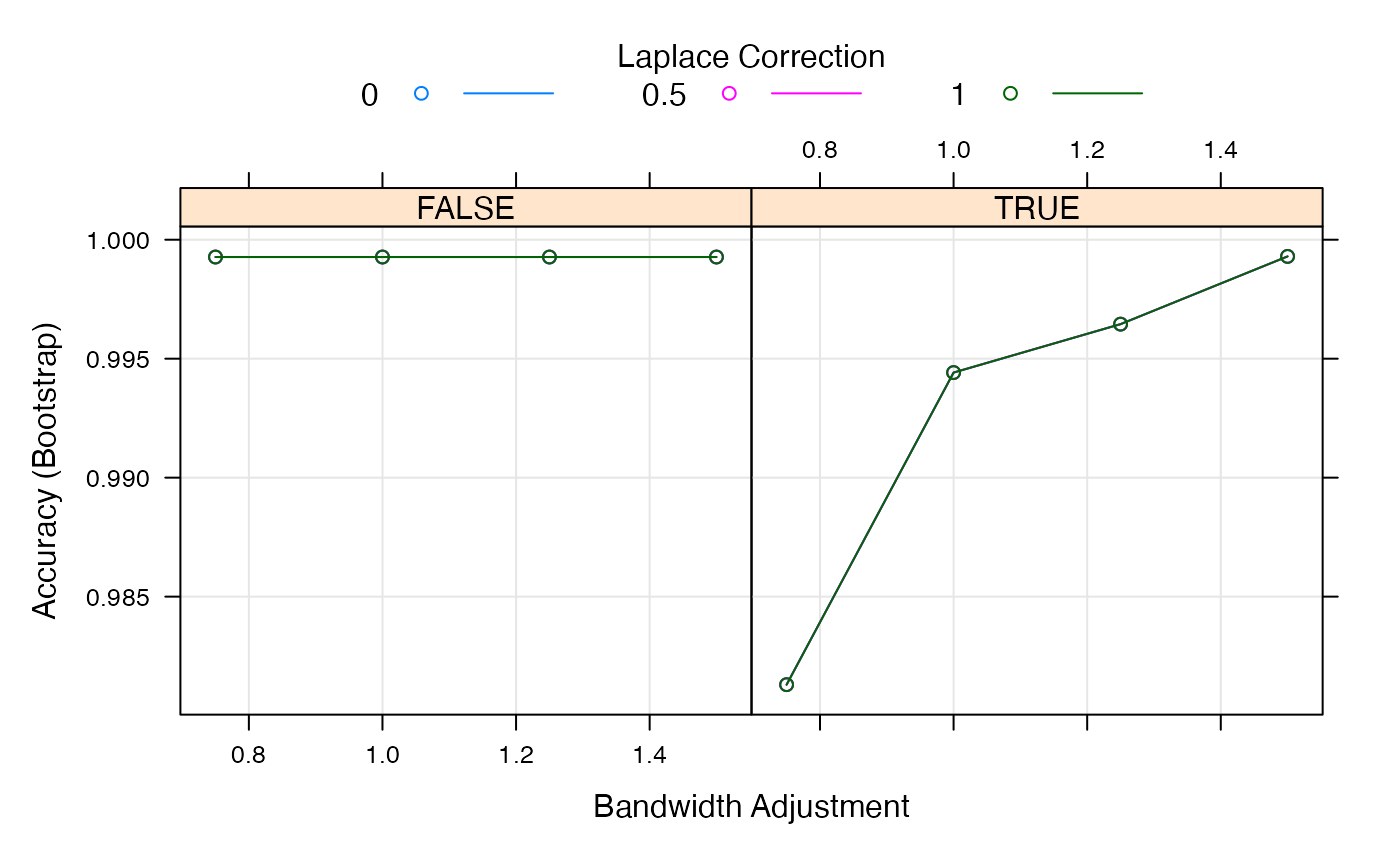
## [1] setosa setosa setosa setosa setosa setosa
## Levels: setosa versicolor virginicaOverall, the code showcases the steps involved in defining a tuning grid, performing model training with parameter tuning, visualizing the tuning process, and applying the trained model for classification using the Naive Bayes algorithm within the caret package in R.
Naive Bayes in nproc
The nproc package
provides Neyman-Pearson (NP) classification algorithms and NP receiver
operating characteristic (NP-ROC) curves, and it can be used in
conjunction with the naivebayes package. By incorporating
the naivebayes package within the NP classification
framework, users can leverage the power of the naive Bayes algorithm for
binary classification tasks while controlling type I and type II errors
effectively. This integration allows practitioners to apply the NP
paradigm to the naive Bayes classifier and enhance its performance in
various application domains.
# Example usage with nproc and naivebayes packages
# install.packages("nproc")
library(nproc)
library(naivebayes)
# Simulate data
set.seed(2550)
n <- 1000
x <- matrix(rnorm(n * 2), n, 2)
c <- 1 + 3 * x[ ,1]
y <- rbinom(n, 1, 1 / (1 + exp(-c)))
xtest <- matrix(rnorm(n * 2), n, 2)
ctest <- 1 + 3 * xtest[,1]
ytest <- rbinom(n, 1, 1 / (1 + exp(-ctest)))
# Train Naive Bayes classifier such that the type I error alpha is no bigger than 5%
naive_bayes_via_nproc <- npc(x, y, method = "nb", alpha = 0.05)
## Recover the "naive_bayes" object
# naive_bayes_via_nproc$fits[[1]]$fit
# Classification
nb_pred <- predict(naive_bayes_via_nproc, xtest)
# head(nb_pred$pred.label)
# Obtain various measures
accuracy <- mean(nb_pred$pred.label == ytest)
ind0 <- which(ytest == 0)
ind1 <- which(ytest == 1)
typeI <- mean(nb_pred$pred.label[ind0] != ytest[ind0]) # type I error on test set
typeII <- mean(nb_pred$pred.label[ind1] != ytest[ind1]) # type II error on test set
cat(" Overall Accuracy: ", accuracy,"\n",
"Type I error: ", typeI, "\n",
"Type II error: ", typeII, "\n")## Overall Accuracy: 0.689
## Type I error: 0.02590674
## Type II error: 0.490228The provided code demonstrates the usage of the nproc
and naivebayes packages in R.
First, the necessary packages are loaded. Then, synthetic data is
generated using the set.seed() function to ensure
reproducibility. The data consists of two independent variables
x and a binary response variable y. A test
dataset xtest is also generated for evaluation.
Next, the naive Bayes classifier is applied using the
npc() function from the nproc package, with
the method set to "nb" for naive Bayes. The
alpha parameter is set to 0.05, defining a
desirable upper bound on type I error.
After training the classifier, predictions are made on the test
dataset using the predict() function. The accuracy of the
predictions is calculated by comparing the predicted labels
nb_pred$pred.label with the true labels
ytest.
Additionally, type I and type II errors are computed by comparing the predicted labels with the true labels separately for class 0 and class 1 instances. The type I error corresponds to misclassifying class 0 observations as class 1, while the type II error represents misclassifying class 1 observations as class 0.
Finally, the overall accuracy, type I error, and type II error are
printed to the console using the cat() function.
The code demonstrates how to leverage the nproc and
naivebayes packages to apply the Neyman-Pearson
classification paradigm1 to the naive Bayes classifier and evaluate
its performance in terms of accuracy and error rates.