
Non-Parametric Naive Bayes Classifier
nonparametric_naive_bayes.Rdnonparametric_naive_bayes is used to fit the Non-Parametric Naive Bayes model in which all class conditional distributions are non-parametrically estimated using kernel density estimator and are assumed to be independent.
Arguments
- x
matrix with metric predictors (only numeric matrix accepted).
- y
class vector (character/factor/logical).
- prior
vector with prior probabilities of the classes. If unspecified, the class proportions for the training set are used. If present, the probabilities should be specified in the order of the factor levels.
- ...
other parameters to
density(for instanceadjust,kernelorbw).
Value
nonparametric_naive_bayes returns an object of class "nonparametric_naive_bayes" which is a list with following components:
- data
list with two components:
x(matrix with predictors) andy(class variable).- levels
character vector with values of the class variable.
- dens
nested list containing
densityobjects for each feature and class.- prior
numeric vector with prior probabilities.
- call
the call that produced this object.
Details
This is a specialized version of the Naive Bayes classifier, in which all features take on real values (numeric/integer) and class conditional probabilities are estimated in a non-parametric way with the kernel density estimator (KDE). By default Gaussian kernel is used and the smoothing bandwidth is selected according to the Silverman's 'rule of thumb'. For more details, please see the references and the documentation of density and bw.nrd0.
The Non-Parametric Naive Bayes is available in both, naive_bayes() and nonparametric_naive_bayes(). The latter does not provide a substantial speed up over the general naive_bayes() function but it is meant to be more transparent and user friendly.
The nonparametric_naive_bayes and naive_bayes() are equivalent when the latter is used with usekernel = TRUE and usepoisson = FALSE; and a matrix/data.frame contains only numeric variables.
The missing values (NAs) are omitted during the estimation process. Also, the corresponding predict function excludes all NAs from the calculation of posterior probabilities (an informative warning is always given).
References
Silverman, B. W. (1986). Density Estimation for Statistics and Data Analysis. Chapman & Hall.
Author
Michal Majka, michalmajka@hotmail.com
Examples
# library(naivebayes)
data(iris)
y <- iris[[5]]
M <- as.matrix(iris[-5])
### Train the Non-Parametric Naive Bayes
nnb <- nonparametric_naive_bayes(x = M, y = y)
summary(nnb)
#>
#> ========================== Nonparametric Naive Bayes ===========================
#>
#> - Call: nonparametric_naive_bayes(x = M, y = y)
#> - Classes: 3
#> - Samples: 150
#> - Features: 4
#> - Prior probabilities:
#> - setosa: 0.3333
#> - versicolor: 0.3333
#> - virginica: 0.3333
#>
#> --------------------------------------------------------------------------------
head(predict(nnb, newdata = M, type = "prob"))
#> setosa versicolor virginica
#> [1,] 1 3.009873e-09 8.846394e-11
#> [2,] 1 4.792767e-08 1.329911e-09
#> [3,] 1 1.950981e-08 1.132901e-09
#> [4,] 1 1.129719e-08 6.470675e-10
#> [5,] 1 8.715390e-10 8.467287e-11
#> [6,] 1 3.746571e-09 5.848304e-09
### Equivalent calculation with general naive_bayes function:
nb <- naive_bayes(M, y, usekernel = TRUE)
summary(nb)
#>
#> ================================= Naive Bayes ==================================
#>
#> - Call: naive_bayes.default(x = M, y = y, usekernel = TRUE)
#> - Laplace: 0
#> - Classes: 3
#> - Samples: 150
#> - Features: 4
#> - Conditional distributions:
#> - KDE: 4
#> - Prior probabilities:
#> - setosa: 0.3333
#> - versicolor: 0.3333
#> - virginica: 0.3333
#>
#> --------------------------------------------------------------------------------
head(predict(nb, type = "prob"))
#> setosa versicolor virginica
#> [1,] 1 3.009873e-09 8.846394e-11
#> [2,] 1 4.792767e-08 1.329911e-09
#> [3,] 1 1.950981e-08 1.132901e-09
#> [4,] 1 1.129719e-08 6.470675e-10
#> [5,] 1 8.715390e-10 8.467287e-11
#> [6,] 1 3.746571e-09 5.848304e-09
### Change kernel
nnb_kernel <- nonparametric_naive_bayes(x = M, y = y, kernel = "biweight")
plot(nnb_kernel, 1, prob = "conditional")
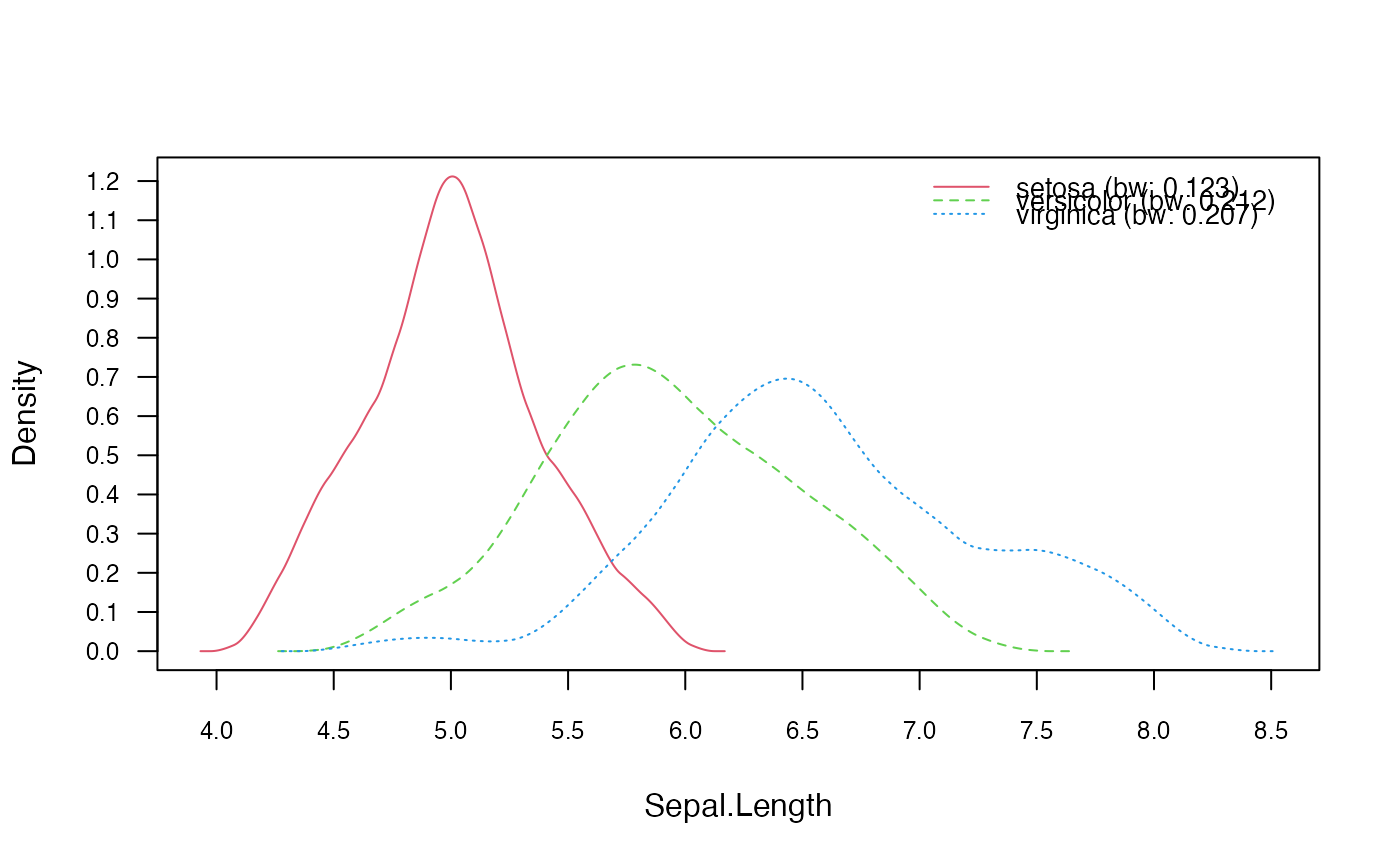 ### Adjust bandwidth
nnb_adjust <- nonparametric_naive_bayes(M, y, adjust = 1.5)
plot(nnb_adjust, 1, prob = "conditional")
### Adjust bandwidth
nnb_adjust <- nonparametric_naive_bayes(M, y, adjust = 1.5)
plot(nnb_adjust, 1, prob = "conditional")
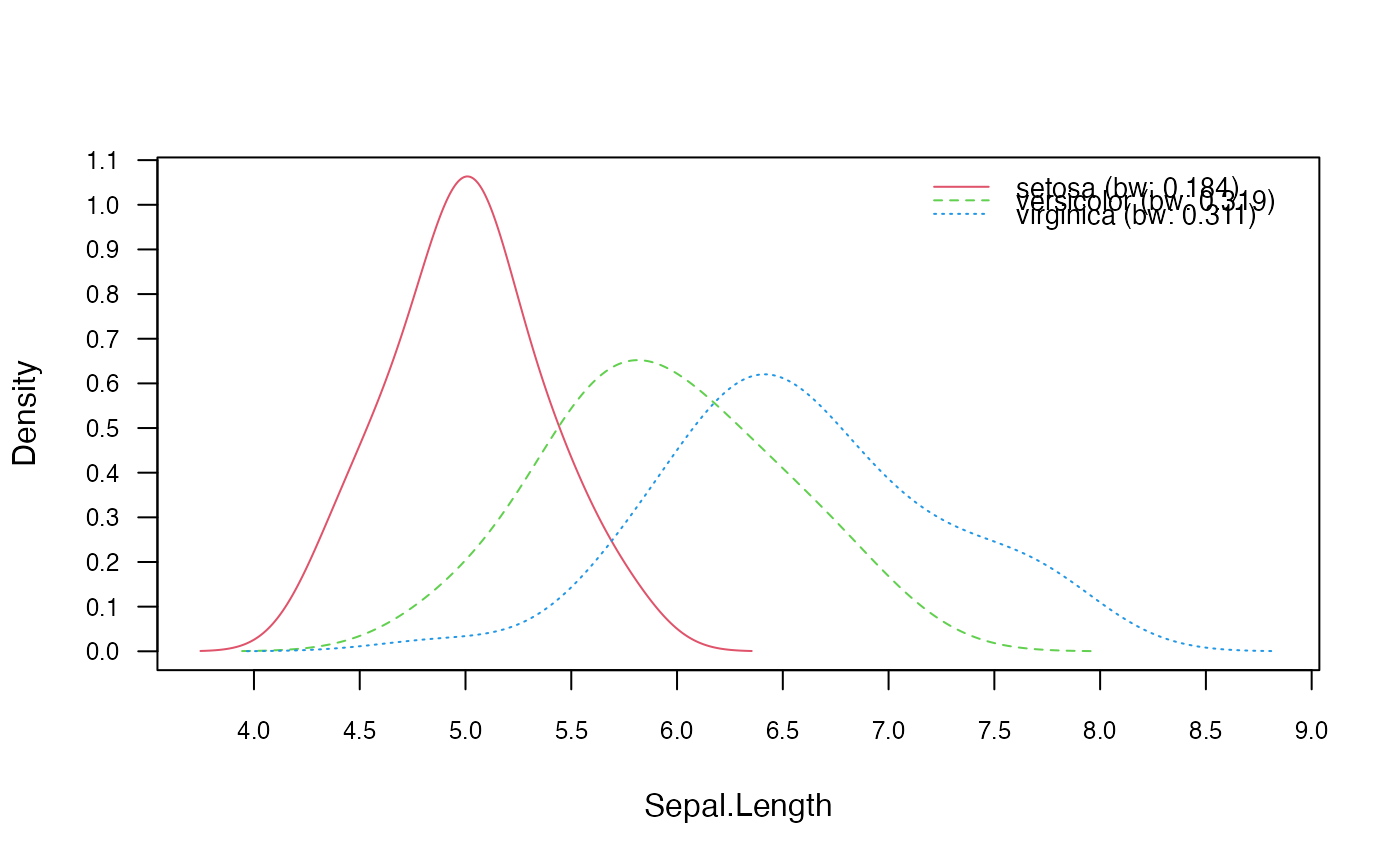 ### Change bandwidth selector
nnb_bw <- nonparametric_naive_bayes(M, y, bw = "SJ")
plot(nnb_bw, 1, prob = "conditional")
### Change bandwidth selector
nnb_bw <- nonparametric_naive_bayes(M, y, bw = "SJ")
plot(nnb_bw, 1, prob = "conditional")
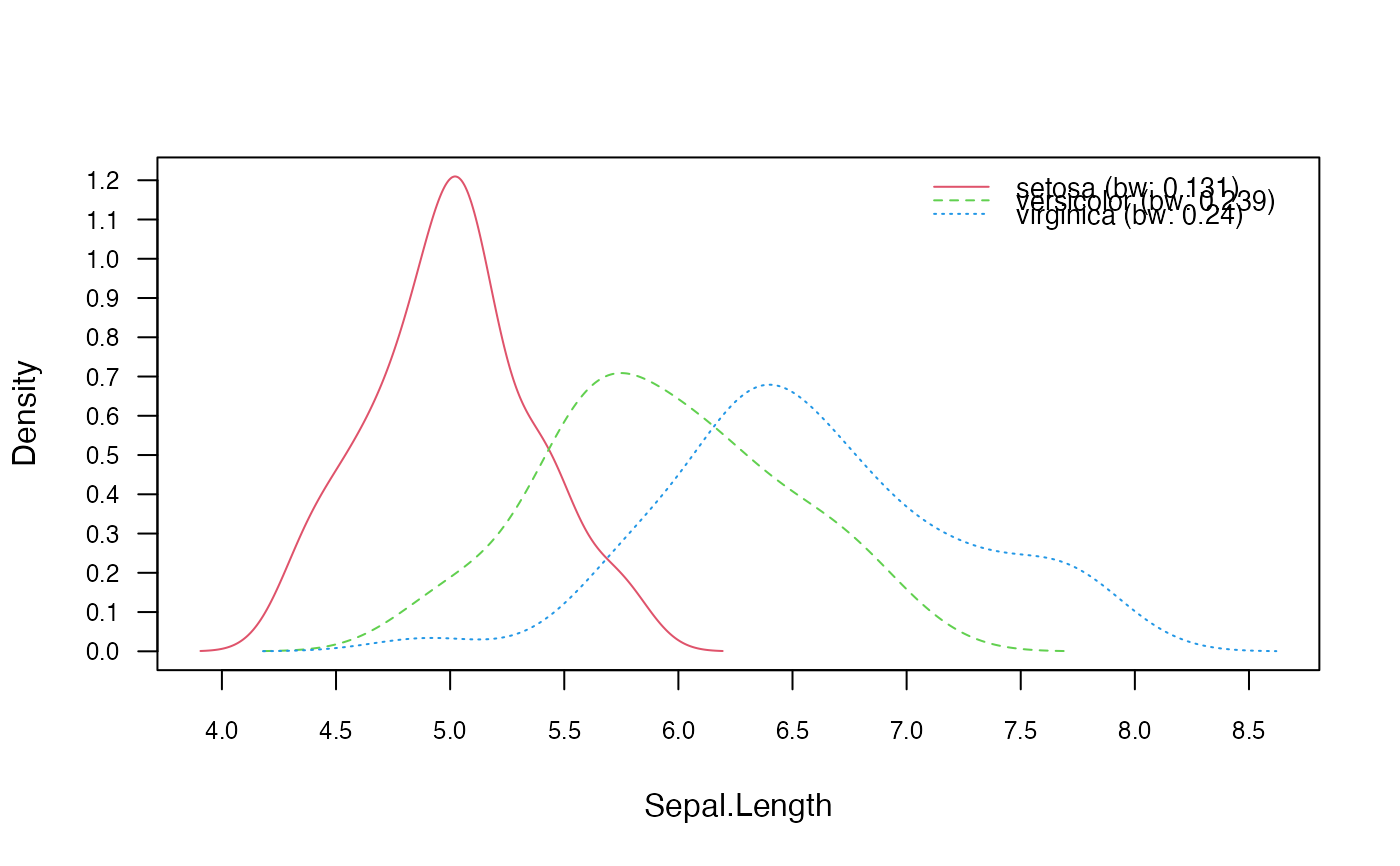 ### Obtain tables with conditional densities
# tables(nnb, which = 1)
### Obtain tables with conditional densities
# tables(nnb, which = 1)